How BostonGene Utilized TabPFN to Identify Immune System Profiles

Background
BostonGene, a pioneer in precision medicine, is leveraging TabPFN in its analysis of multi-modal data from genomic and immune system profiling to enhance success for drug developers and improve patient outcomes. In the May 2024 edition of Cancer Cell, BostonGene demonstrated how immune system profiling of peripheral blood cells can monitor disease states and predict treatment responses in patients with advanced cancers.
By integrating Prior Labs' tabular foundation model, TabPFN, with its machine learning-based immune system profiling platform, BostonGene was able to distinguish cancer patients from healthy individuals with 90% accuracy using only data from the patient’s peripheral blood immune cells. Importantly, this method does not require any analysis of the patient’s tumor & overcame challenges of data availability.
This TabPFN-based classifier subsequently informed the development of a comprehensive immune system model capable of predicting responses to cancer immunotherapy. This enables oncologists to personalize immunotherapy regimens more effectively.
The Challenge
The immune system holds immense information about an individual’s overall health, including how the body is responding to a disease like cancer and how it is positioned to respond in the future. Making sense of this data to identify clinically useful biomarkers requires sophisticated extraction techniques and cutting-edge analytical tools. However, the testing methods used to generate these critical datasets presented several key challenges:
- Small Datasets: Despite the complexity of collecting immune system profiles from peripheral blood samples, each dataset remains relatively small. Traditional ML models are prone to overfitting on such limited data, making it difficult to generate reliable insights.
- Time-Consuming Hyperparameter Tuning: With few solutions designed for small, tabular datasets, BostonGene’s data scientists had to manually test and refine multiple models, consuming valuable time and resources while risking performance limitations.
- High-Dimensional Cell Profiling Multiparameter flow cytometry yields an expansive set of cell phenotypes—over 650 immune cell types and activation states—making it challenging to identify which features are most closely associated with disease states and treatment responses.
Michael Goldberg, PhD, VP R&D at BostonGene
“Accurately classifying cancer patients and healthy individuals based on the distribution of immune cells in the peripheral blood was a remarkably challenging task — TabPFN made it a reality”
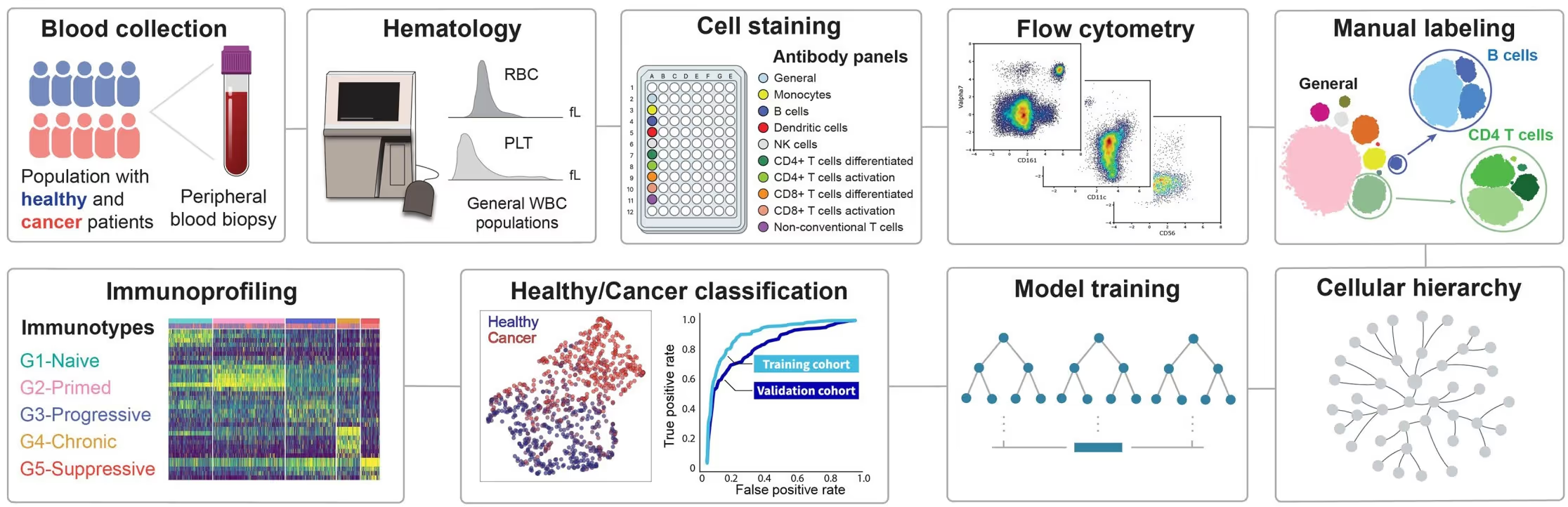
The Approach
BostonGene’s research team devised an end-to-end pipeline pairing multi-parameter flow cytometry with advanced ML:
- Data Acquisition and Cell Typing: Detailed immune cell profiles were collected from both healthy donors and cancer patients, identifying over 650 immune cell types and activation states.
- Feature Selection: From these data, the team identified 20 critical immune cell populations that most effectively distinguished cancer patients from healthy donors.
- Integration of TabPFN: TabPFN specifically addresses challenges associated with small datasets by using meta-learning to prevent overfitting. Additionally, it eliminates the need for extensive hyperparameter tuning. As a result:
90% reduction in model development time: TabPFN autonomously optimizes model settings, cutting model development time by 90%. Improved generalization: As a pre-trained foundation model, TabPFN effectively learns from limited samples, reducing overfitting and boosting predictive accuracy. The TabPFN-based classifier achieved a ROC-AUC of 0.91, significantly outperforming conventional models.4. Cluster Analysis: By analyzing TabPFN’s outputs, BostonGene identified distinct immune cell clusters associated with patient responses, validating crucial hypotheses about how different patient immune profiles correlate with immunotherapy success.
Breakthrough Results
- Unmatched Accuracy in Limited Data Scenarios: By automating model selection and tuning, TabPFN delivered high sensitivity and specificity, effectively distinguishing cancer patients from healthy donors without overfitting—even with minimal data.
- 90% Faster Workflow: By eliminating manual hyperparameter tuning, TabPFN enabled BostonGene’s data scientists to work 90% faster, allowing them to bring insights to clinical stakeholders more efficiently.
- Clinical Impact: With more precise immune system profiling, BostonGene’s platform provides critical prognostic intelligence for treatment response, enabling oncologists to personalize immunotherapy regimens more effectively.
Arseniy Sokolov, former Data Scientist at BostonGene
“TabPFN eliminated the tedious hyperparameter tuning process, allowing me to focus on core scientific questions instead of technical optimization. It accelerated model development by 90% while achieving excellent classification performance.”
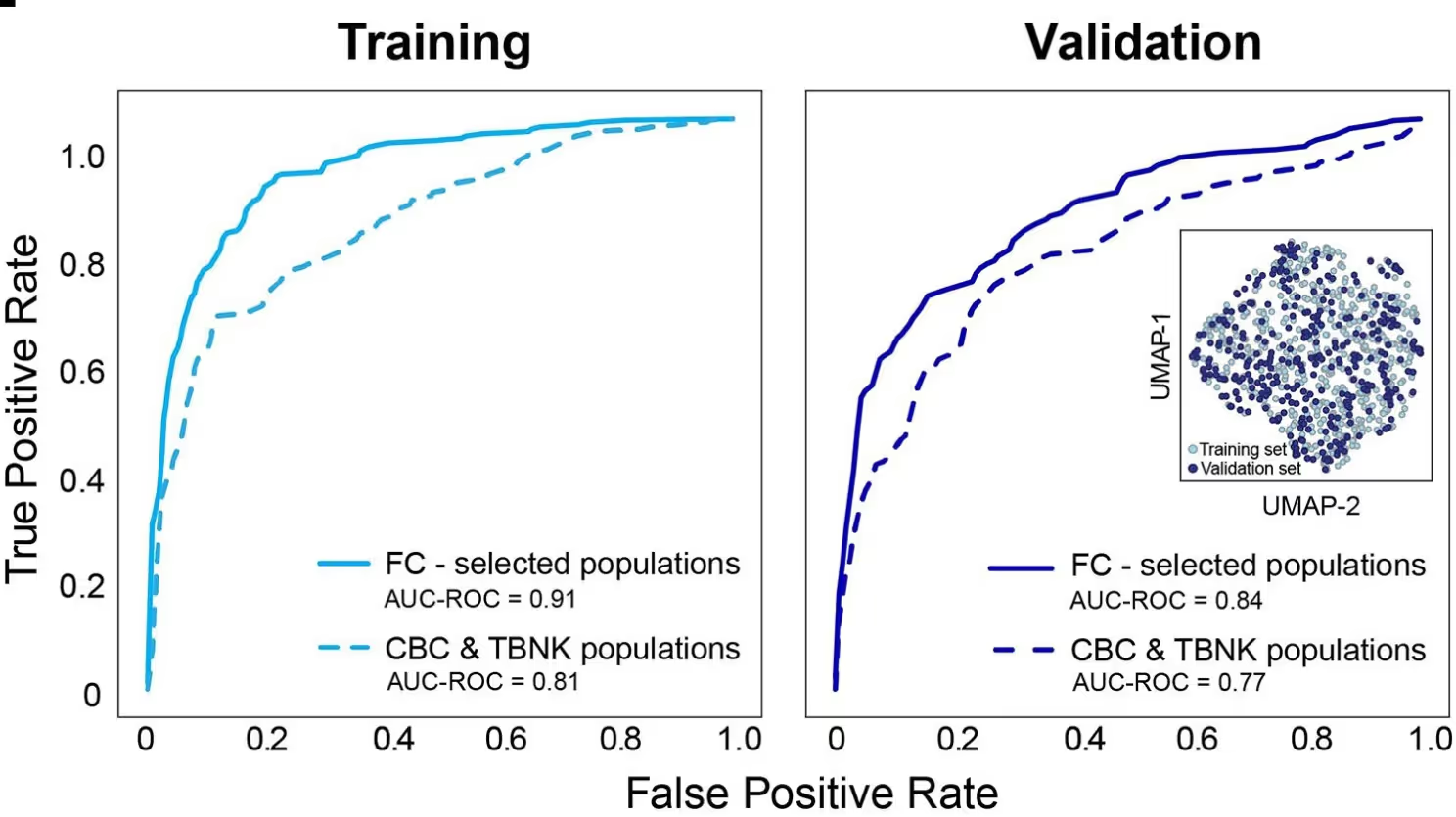
Conclusion
BostonGene’s implementation of TabPFN represents a significant advancement, demonstrating how peripheral blood immune system profiling can identify disease states and predict treatment responses.
By integrating advanced machine learning into its immune system profiling platform, BostonGene overcame the challenges of limited data availability and achieved breakthroughs in patient stratification and analysis speed. This case study underscores the transformative potential of TabPFN in clinical applications, highlighting its value as a powerful tool in biomarker discovery in immuno-oncology–enhancing both predictive accuracy and operational efficiency.
